
ElemBio Catalyst enables customers to generate and share FASTQ files in ElemBio Cloud. We are excited to extend these capabilities with highly accurate and efficient secondary analysis workflows for whole genome sequencing and enrichment panel applications. For those interested in automated secondary analysis without the need for local compute or expertise in cloud integration, we now offer automated generation of BAM and VCF files using your choice of NVIDIA® Parabricks® or Sentieon DNAScope workflows. Sentieon offers accurate, fast, and cost-effective tools to accelerate BWA to align reads and call variants using DNAScope, coupled with AVITI-specific machine learning models to increase accuracy. Parabricks leverages GPUs to accelerate BWA alignment and Google DeepVariant variant calling, trained on various platforms including AVITI. These workflows have been optimized by Element for accuracy, speed, and cost. Some benefits of using these workflows include:
- Powerful alignment, variant calling, and reporting tools from industry leaders NVIDIA Parabricks, Sentieon DNAScope, and Google DeepVariant
- Automatically use Element-specific machine learning models designed for AVITI
- Variant calling models trained with Cloudbreak, Cloudbreak Freestyle, and Trinity data from AVITI
- Ease of use and powerful automation, for experts and non-experts
- Scalability to any number of samples (burst capacity built-in)
Workflow Highlights
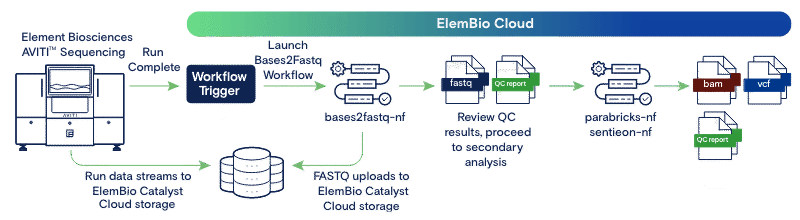
ElemBio Catalyst x NVIDIA
The Parabricks Germline DNA Sequencing flow integrates NVIDIA’s Parabricks fq2bam (BWA alignment, co-ordinate sorting, marking duplicates, and Base Quality Score Recalibration) and Parabricks accelerated DeepVariant to deliver a comprehensive whole genome or enrichment analysis. Parabricks leverages GPU power to accelerate genomic tasks like read alignment and variant calling, significantly reducing processing time and cost while providing equivalent results. Google DeepVariant uses deep learning to enhance variant calling accuracy by interpreting sequencing data through a neural network model.
Highlights
- GPU-accelerated computing for faster analysis
- Support for Whole Genome and enrichment analysis across multiple panel
- Process large sample datasets quickly
- Simplified workflow for a streamlined analysis
- Excellent variant calling accuracy across various coverage levels benchmarked for AVITI data
ElemBio Catalyst x Sentieon
The Sentieon Germline DNA Sequencing flow integrates Sentieon's DNAscope for whole genome and enrichment analysis. DNAscope integrates GATK’s HaplotypeCaller preprocessing and assembly methods with a machine-learned genotyping model. Benchmark comparisons between DNAscope and DNAseq (Sentieon’s GATK-compatible germline variant calling pipeline) demonstrate that DNAscope provides superior SNP and indel accuracy while reducing computational costs. Sentieon technology delivers high accuracy and ease of use, with a significant boost in processing speed.
Highlights
- Sentieon licensing is managed by Element and included with every ElemBio Catalyst subscription
- Support for Whole Genome and enrichment analysis across multiple panels
- Cost efficient CPU scaling of cloud compute
- Simplified workflow leveraging Sentieon CLI for a streamlined analysis
- Highly accurate variant detection, especially in challenging regions, utilizing machine learning (see app note)