MicroRNA Sequencing
MicroRNA (miRNA) sequencing assesses small, noncoding RNA molecules to grant researchers insight into key biological functions for profiling and discovery applications. The Element AVITI™ System delivers consistent, accurate data at a price point that scales to serve a 2 x 75 read length and output of ~5–10 million reads per sample.
AVITI also offers the flexibility to explore both existing and novel miRNA.
- High sequencing quality with high recovery
- Consistent correlation and expression of miRNA
- Ability to scale experiments for small inserts and lower output
- Compatibility with third-party library preps via the Adept™ Workflow or Cloudbreak Freestyle™ sequencing chemistry
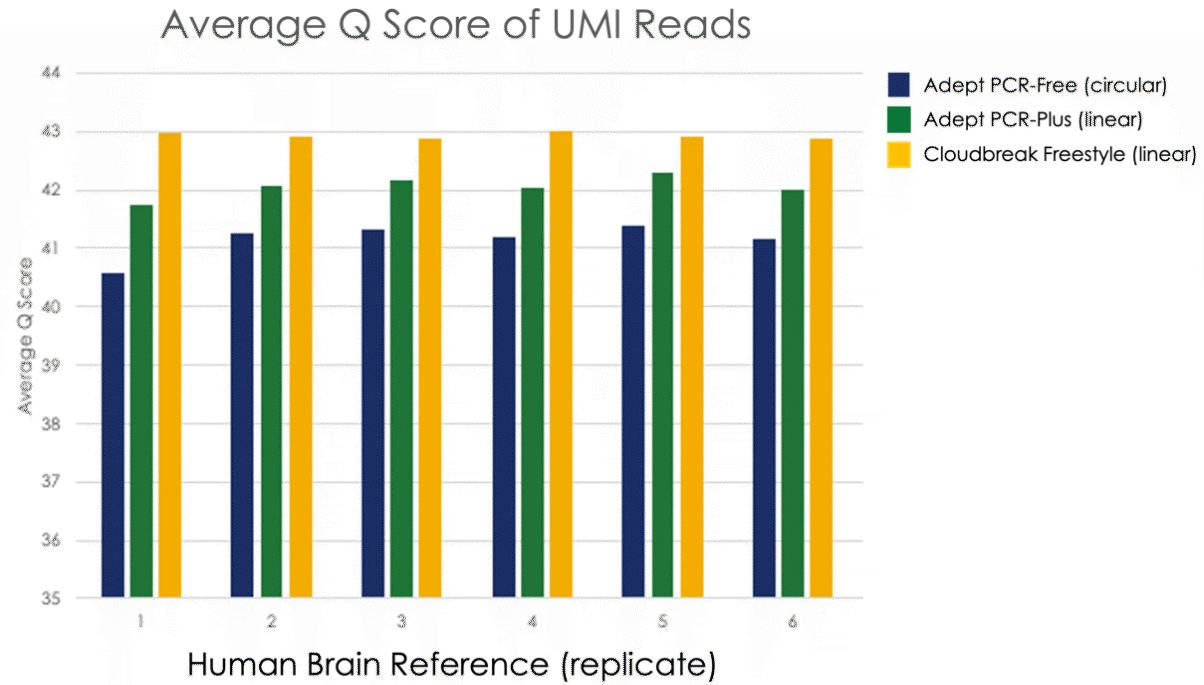
Figure 1. Unique molecular identifier (UMI) reads from six replicate libraries prepared from Human Brain Reference Total (HBR) RNA and sequenced on AVITI deliver high quality scores (Q-scores) of 41–43.
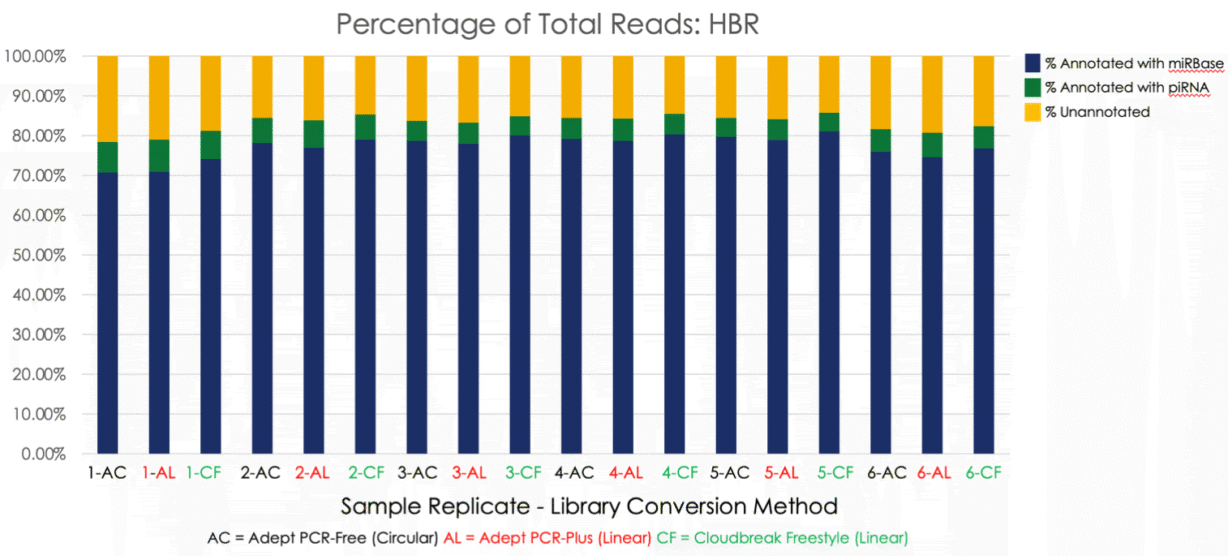
Figure 2. miRBase and piRNA annotate a high percentage of mapped mRNA molecules across six replicate libraries prepared from Human Brain Reference (HBR) RNA. Each method of converting third party libraries for sequencing on AVITI, Adept Rapid PCR-Free, Adept Rapid PCR-Plus, and Cloudbreak Freestyle deliver similar results.