What better way to start the year than attending our first PAG, the largest international gathering of plant and animal genomics scientists in the world, held right in our backyard in San Diego, California. We were excited to see just how many attendees came eager to talk to us about Element’s mission to disrupt genomics with high-quality, low-cost DNA sequencing data on the AVITI. Coming on the heels of our $200 Genome Program announcement at the JP Morgan Healthcare conference, PAG participants were ready to learn how they, too, could benefit from our new throughput-based pricing model to further bring down the cost of sequencing for the applications most important to them, including whole genome sequencing, genotyping by sequencing, low-pass genome sequencing for genotype selection, and RNA sequencing.

In addition to instrument demos, PAG attendees had multiple options for seeing AVITI system data on the scientific program. On Monday, Alison Devault from Daicel Arbor Biosciences presented a poster “A universal targeted sequencing system for any high-throughput sequencing platform,” where she described the myBaits® enrichment system, which is compatible with virtually any input nucleic acid and sequencing platform. The poster featured data showing that for five distinct assay types, the AVITI returned comparable or better library representation, reads on-target, and locus retrieval as NovaSeq despite highly variable length distributions and target compositions."
That same afternoon, Jacky Carnahan from Neogen Genomics presented a poster “Low pass sequencing in animals: Element Biosciences AVITI as a useful platform.” The poster featured a three-way collaboration between Neogen, Gencove and Element, describing an end-to-end solution for low pass sequencing and genotype imputation. The genotyping calls were highly consistent with results from a production-scale DNA sequencing instrument, but lower duplication rates yielded higher effective coverage. Additionally, the reagent cost per Gb was comparable to the higher throughput system, providing a cost-competitive path to potentially faster turnaround times at lower sample volumes.
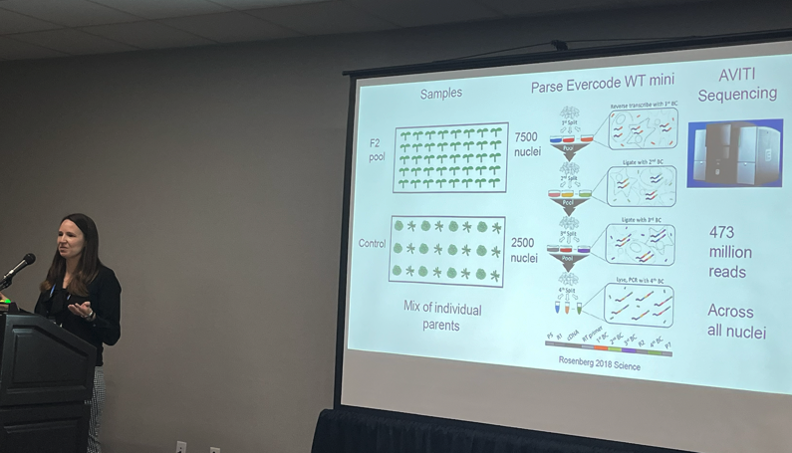
Finally, Beth Rowan presented preliminary data on her methods development project, “Evaluation of Pooled and Single-Nucleus Sequencing Approaches for the Study of Recombinant Populations and the Generation of Genetic Maps.” The overall goal of her study is to use CRISPR to alter genes involved in the regulation of meiotic recombination and for that she needed to develop a rapid method for detecting the frequency and locations of meiotic crossovers. Pooling and sequencing recombinants offers an efficient, high-throughput readout for assessing changes in crossing-over events after genome editing, with TELL-seq linked read sequencing and single cell RNA sequencing as two options for finding crossover breakpoints in recombinant populations. Dr. Rowan reported that the TELL-seq method looks both promising and cost effective in a low coverage test run, and she plans to scale up her pilot, potentially on the AVITI. She also shared preliminary data on single cell RNA-seq using Parse and sequencing on a shared AVITI flow cell. While this approach will require further optimization to reduce rRNA representation in the data, Dr. Rowan noted that “The AVITI data was high quality, and I can recommend the system as a practical and cost-effective option for methods development pilot studies.”
Looking to learn more about how an AVITI system can accelerate your plant and animal research? Come visit us at AGBT 2023 or AGBT Ag 2023, or read our blog post on single cell RNA sequencing on the AVITI system.