What is hybrid capture?
Hybrid capture is an essential biology technique used across many genomic applications, from cancer research and disease biology to microbial genomics, agriculture, and exome sequencing1-5. The hybrid capture process enables you to focus your sequencing efforts on specific regions of the genome, maximizing the sequencing reads applied to your regions of interest so you don’t waste reads.
Unlike whole genome sequencing, hybrid capture hybridizes your library to a set of probes that are specific to your application or organism. The probe set can be large or small, spanning just a few genes to an entire exome. This provides higher coverage for those regions of interest compared to a standard whole genome sequencing prep, allowing more samples to be sequenced on a single flow cell and reducing overall sequencing costs.
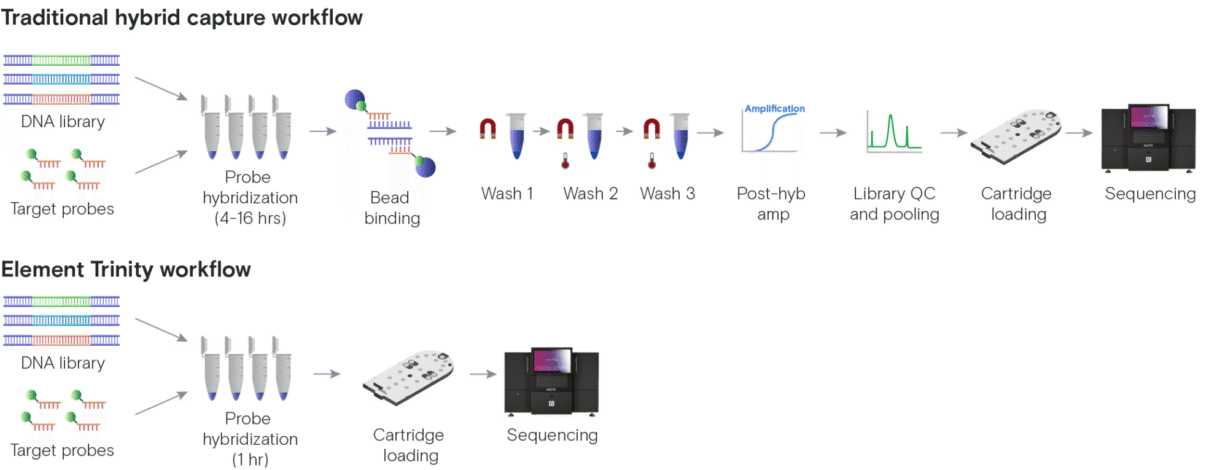
However, hybrid capture protocols can be lengthy and complex, and are often split into multiple workdays to complete2,5. After hybridization, traditional methods use magnetic beads and a series of temperature-controlled wash steps to isolate the hybridized material and remove non-specific material. While these steps help achieve a high on-target rate for your panel, they are also time-consuming and can lead to a large loss of DNA. Due to this loss, these workflows usually require PCR amplification just to generate enough material for sequencing. For some, the cost savings are worth the complicated manual procedures and extra hours in the lab.
In our recent pre-print publication about Trinity™, we show that you don’t have to compromise: the Trinity workflow on an AVITI™ system reduces the hands-on and total time required for a hybrid capture workflow, all while maintaining or exceeding the performance of traditional hybrid capture.
How is Trinity different?
When the Element team first thought about how to improve hybrid capture workflows, we knew we wanted to decrease and simplify the manual steps in the process. The team developed a new flow cell surface and new reagents capable of capturing the library fragments bound to the probes, successfully eliminating all of the manual post-hybridization steps from the workflow.
Trinity still hybridizes the library to the same panel of probes as the traditional workflow, but the hybridized library can then be loaded directly onto the instrument. This means there is no bead capture, no time-consuming and finicky manual wash steps, and no post-capture PCR amplification needed!
Faster, easier prep with exceptional results
Eliminating manual post-hybridization steps provides a huge time savings in the hybrid capture workflow. While the traditional workflow had to be broken up into at least two days to complete all steps prior to sequencing run set up, the entire Trinity workflow, from library preparation through sequencing run prep, can be completed in as few as five hours using a 1-hour hybridization step.
To further validate the robustness of the technology, we tested multiple capture panels down to 700kb and found that Trinity’s performance was consistent for multiple vendors and across panel sizes. With the reduction in post-hybridization PCR and washing, we generally saw improved duplicate rates and library complexity, higher mean target coverage, and better variant calling benchmarking. We also present a unique opportunity to eliminate PCR entirely from the hybrid capture process and sequence a completely PCR-free exome with Trinity.
Dive into the details by reading our pre-print here: A simplified hybrid capture approach retains high specificity and enables PCR-free workflow | bioRxiv.
References
- Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A 106, 19096-19101 (2009).
- Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27, 182-189 (2009).
- Kim, D.W., Nam, S.H., Kim, R.N., Choi, S.H. & Park, H.S. Whole human exome capture for high-throughput sequencing. Genome 53, 568-574 (2010).
- Bansal, V., Tewhey, R., Leproust, E.M. & Schork, N.J. Efficient and cost effective population resequencing by pooling and in-solution hybridization. PLoS One 6, e18353 (2011).
- Xiao, W. et al. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat Biotechnol 39, 1141-1150 (2021).