With the steadily decreasing cost of sequencing, low pass whole genome sequencing with imputation is gaining traction in agriculture as a method of genotyping. In this Element 101 blog series post, we dig into how low pass WGS works, what it can offer beyond targeted genotyping options like PCR and microarrays, and how the Element AVITI™ system can help even moderate throughput labs make the leap to this high information, cost-effective method.

Balancing information density and cost per sample
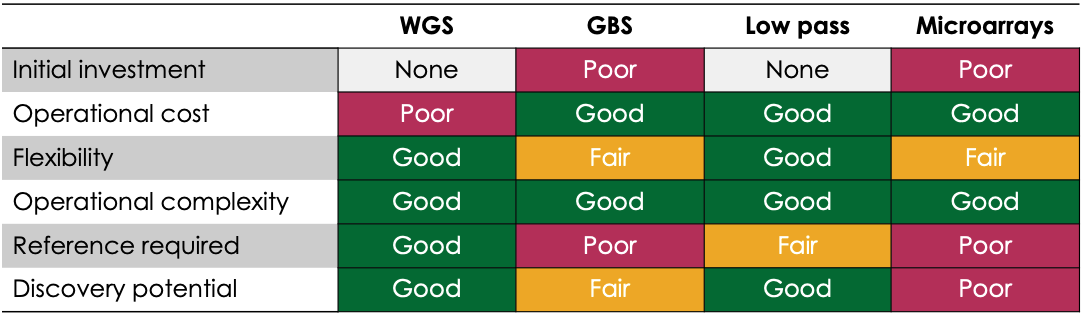
In an ideal world, genotyping could be done using 30-fold coverage whole genome sequencing, supplying high confidence data across all possible variants in one easily operationalized assay. However, the high cost per sample combined with the slow turnaround time for whole genome sequencing at the scale required for commercial genotyping makes this impractical. Microarrays have long been an appealing solution for genotyping, as they offer:
- Low cost
- Ease of use
- Scalable number of markers
However, microarrays come with their own compromises:
- Requirement for a reference genome
- High design, startup, and revision costs
- Low potential for variant discovery, as only known targets can be included on microarray chips
- Potentially poor sampling of genetic diversity from non-domesticated or divergent strains
As the cost of genome sequencing has continued to drop, new targeted sequencing-based methods have been developed as ways to improve the discovery power of genotyping methods while still controlling cost per sample. One popular method of genotyping by sequencing (GBS) involves DNA digestion with restriction enzymes, followed by adapter ligation, amplification, and sequencing. Another variation involves highly multiplexed PCR reactions to amplify known markers, followed by amplicon sequencing. Hybridization capture is yet another alternative for focusing sequencing on markers of interest. All methods offer notably improved potential for the discovery of new variants linked to important traits. GBS methods come with their own limitations, though:
- Like microarrays, GBS requires a reference genome and significant investments in assay development and validation.
- The manufacture of probes for target capture GBS requires a large initial investment and economies of scale to achieve favorable pricing.
- The target capture protocol is laborious and requires relatively high DNA input quantities.
Working with GBS service providers can make these methods more accessible, particularly for commonly studied organisms, but outsourcing can increase turnaround times and it may be preferable to maintain internal control of samples.
Low pass genome sequencing paired with imputation has the potential to overcome many of these remaining barriers to the adoption of NGS for genotyping by combining assay simplicity, high information content, and affordable cost per sample. In this method, samples are sequenced to a depth of just 0.4 to 1.0-fold coverage and imputation analysis is employed to backfill missing sequences using prior knowledge of gene variant co-inheritance patterns. While a reference genome is still needed for genotype calling from low pass sequencing data, the advantages over microarrays, in particular, are clear:
- No reagent startup costs related to assay development or redesign.
- Operational simplicity, with commercially available options for high throughput library preparation and data analysis support.
- Markers across the entire genome can be leveraged for simultaneous trait selection and variant discovery.
- Improved statistical power for genome-wide association studies compared to microarray assays1
- As more samples are sequenced and added to a low-pass WGS database, the potential for discovery of novel variants increases, including through re-analysis of archived data.
Low pass WGS on the Element AVITI system
Low pass genome sequencing has the strong potential to simply and accelerate breeding programs, however it is only now becoming practical from a cost standpoint. In order to achieve favorable pricing, users must typically be able to batch very large numbers of samples for sequencing on ultra-high throughput systems. This has been achievable for large breeding operations where samples can be run centrally, or by smaller operations that are able to outsource. Now, however, the Element AVITI DNA sequencing platform offers an alternative.
The AVITI is unique as a highly accurate, mid-throughput DNA sequencer with cost per gigabase comparable to production scale systems. At ~$5 per gigabase, low pass sequencing is affordable even with dramatically fewer samples to run. The AVITI also offers technical advantages, including negligible index hopping and high effective coverage stemming from a low duplication rate.2
Customers with mid-level sample throughput can affordably run samples in-house, reducing turnaround times while maintaining possession of valuable materials and data. For large-scale operations, the AVITI opens the door to decentralization. Turnaround times can be reduced by having genotyping facilities at or near globally dispersed production sites to take advantage of year-round growing cycles. With throughput based pricing available through Element’s $200 Genome Program, sequencing costs can be further reduced to as low as $2 per gigabase.
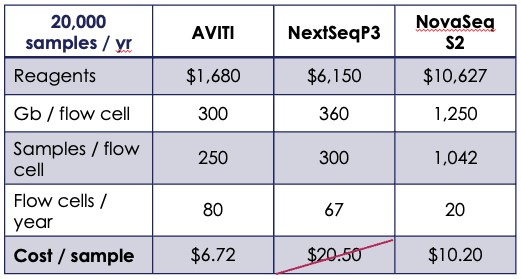
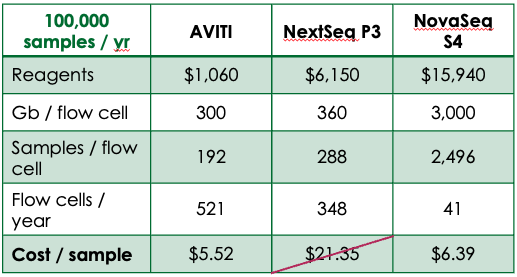
While switching over to a new genotyping method is a significant lift, the AVITI presents a new route for breeding operations of all scales to affordably increase the power of their genotyping pipeline. The implementation of a new workflow can be simplified by working with validated Element ecosystem partners for both library prep and imputation analysis. Multiple options exist for high-throughput library preparation for the AVITI, including purePlex from seqWell, a Tn5 transposase-based system that facilitates even pooling of libraries at scale. For analysis, Gencove offers an enterprise analytics platform for low pass imputation that simplifies genotype calling and report generation.
To learn more about whether the AVITI is right for your lab, talk to an Element scientist by filling in the form on this page, or contact us here.
References
1. Li et. al. (2021). Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays. Genome Res. 31(4):529-537. doi: 10.1101/gr.266486.120.
2. Li et. al. (2022). Low-pass sequencing plus imputation using avidite sequencing displays comparable imputation accuracy to sequencing by synthesis while reducing duplicates. BioRxiv. doi: 10.1101/2022.12.07.519512.